Running MLwiN using mlnscript via the R2MLwiN R package on Apple Silicon Macs
Introduction
MLwiN from the Centre for Multilevel Modelling (CMM) at the University of Bristol (disclaimer: where I also work) is a fantastic piece of software (Charlton et al. 2024). The name suggests it only works on Windows, but as we’ll find out this is very much not the case.
However, in the past this was sort of true because to make it work on a Mac (or Linux machine) with an Intel processor one would need to run it using Wine.
More recently, CMM have cleverly made the MLwiN libraries available for other operating systems in a command line version of the program called mlnscript and an accompanying library. The files for macOS are universal binaries which means that they run natively on both Intel and Apple Silicon Macs. Let’s find out how to set this up.1
Setting up mlnscript on an Apple Silicon Mac
Obtain the installer for macOS. See the relevant download page (depending upon whether you are an academic) on the MLwiN website. On the form on the File to download dropdown menu select the mlnscript for MacOS option. This will give you the MLN.dmg installer.
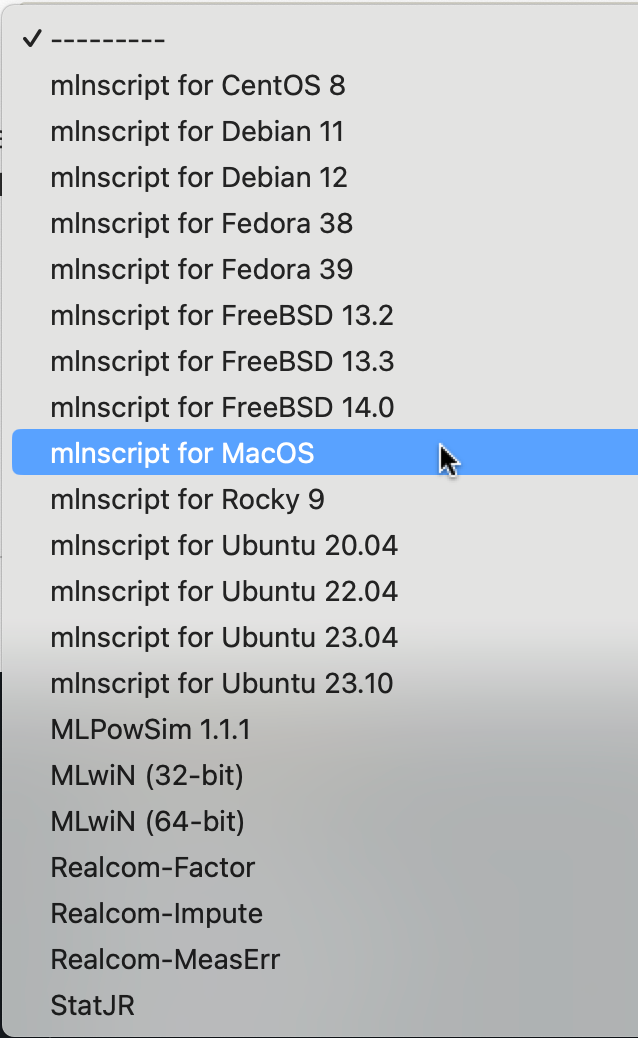
Double click the installer. On macOS it is recommended to install the files into the /opt/mln/ directory, which you will need to create with Admin permissions, or install to another directory if you don’t have Admin permissions. Copy the 2 files mlnscript and libmln.dylib into the /opt/mln (or other) directory.
Once installed we can check that mlnscript and libmln.dylib are universal binaries as follows (we could also use the
filecommand).lipo -archs /opt/mln/mlnscript ## x86_64 arm64Since both architectures are listed in the output this indicates the files are universal binaries. Apple Silicon Macs will use the arm64 architecture.
Now we need to grant the two files permission to run. To do this run the following in your Terminal.
/opt/mln/mlnscript --versionOn first run, this will fail with a pop-up similar to the following.
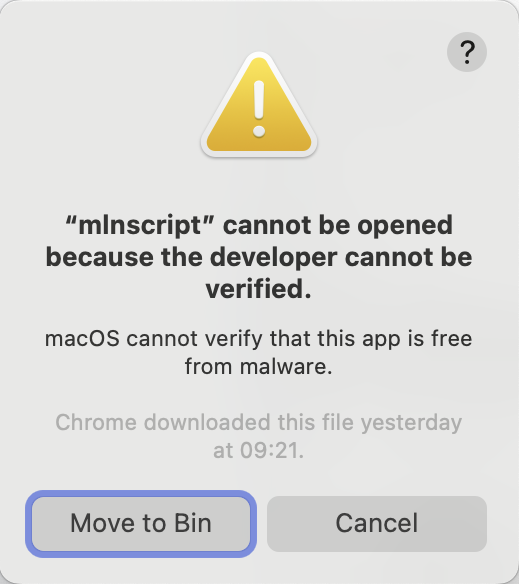
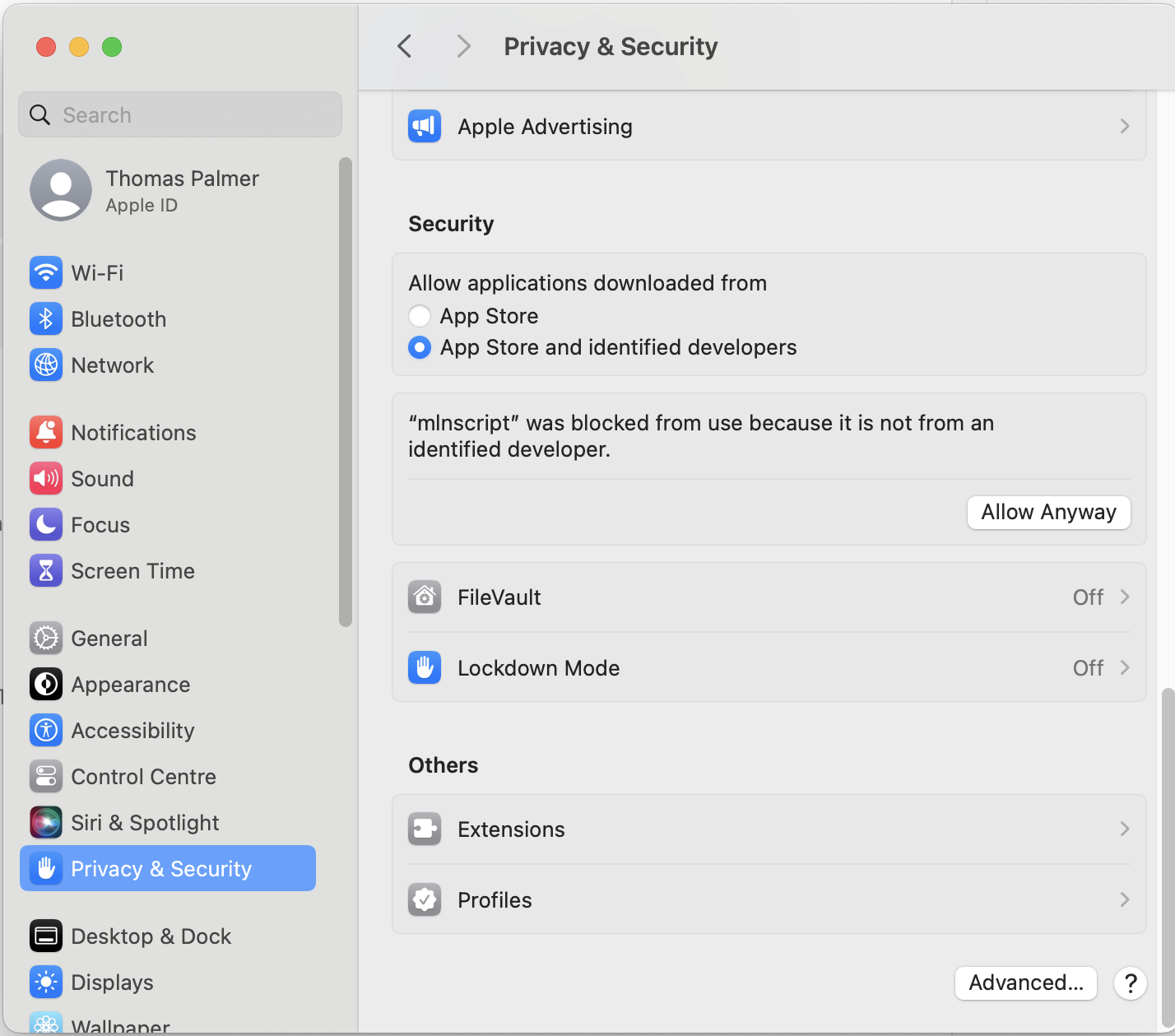
Click Cancel and then go into the System settings | Privacy & Security and scroll down and click Allow Anyway.
Then running the version check command again you may receive another popup in which you click Open.
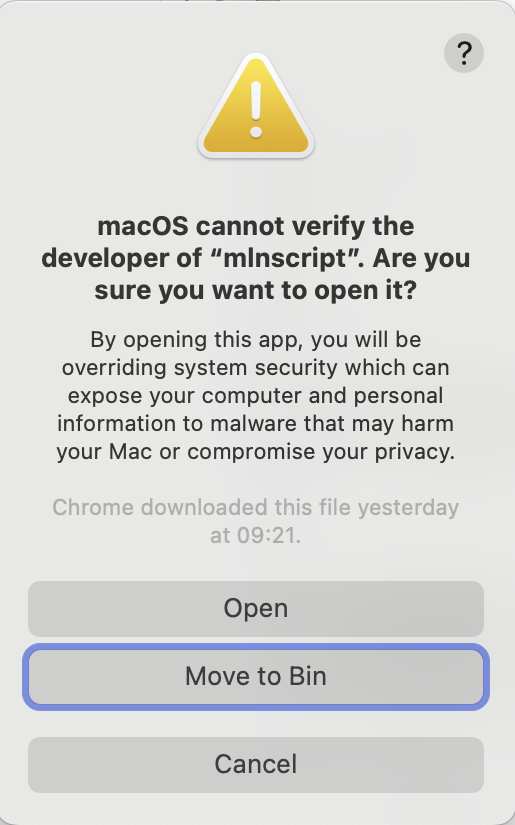
After this the first popup will then appear but about the libmln.dylib file. Again set the security setting to Allow All.
Now running the version check command again you should see the version number – which is currently 3.10.
/opt/mln/mlnscript --version
## 3.10In R we then install the R2MLwiN package from CRAN (Zhang et al. 2016).
install.packages("R2MLwiN")
This completes the setup - phew 😮!
Running a multilevel model
For an example we could run one of the demos in the package, we can list those with the following code.
demo(package = "R2MLwiN")We can run one, for example, let’s fit the random intercept model from the UserGuide02 demo.
library(R2MLwiN)
# if you did not install mlnscript and libmln.dylib in /opt/mln , set:
# options(MLwiN_path = "/path-to/mlnscript")
(mymodel1 <- runMLwiN(normexam ~ 1 + sex + (1 | student), data = tutorial))
#>
#> -*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-
#> MLwiN (version: unknown or >3.09) multilevel model (Normal)
#> Estimation algorithm: IGLS Elapsed time : 0.03s
#> Number of obs: 4059 (from total 4059) The model converged after 3 iterations.
#> Log likelihood: -5727.9
#> Deviance statistic: 11455.7
#> ---------------------------------------------------------------------------------------------------
#> The model formula:
#> normexam ~ 1 + sex + (1 | student)
#> Level 1: student
#> ---------------------------------------------------------------------------------------------------
#> The fixed part estimates:
#> Coef. Std. Err. z Pr(>|z|) [95% Conf. Interval]
#> Intercept -0.14035 0.02463 -5.70 1.209e-08 *** -0.18862 -0.09208
#> sexgirl 0.23367 0.03179 7.35 1.985e-13 *** 0.17136 0.29598
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
#> ---------------------------------------------------------------------------------------------------
#> The random part estimates at the student level:
#> Coef. Std. Err.
#> var_Intercept 0.98454 0.02185
#> -*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-*-We can see the output is in several sections. The first section tells us about how mlnscript, which estimation algorithm it used, hwo long it took to fit the model, and some characteristics of the dataset. The second section tells us about the model, in this case a random intercept model. The third section is the fixed effect estimates and the associated statistical inference for them. The fourth section is the random effect variance estimates.
And we can continue with our multilevel modelling as we like.
Summary
Despite having Win in its name, MLwiN is available as a command line program, mlnscript, which is available on operating systems other than Windows (and indeed with other architectures), including macOS for both Intel and Apple Silicon processors and various Linux and Unix distributions (CentOS, Debian, Fedora, FreeBSD, Rocky, and Ubuntu). This is straightforward to use from R via the R2MLwiN package.